4 Case Study
The loss of uricase during hominid evolution has predisposed humans to hyperuricemia and gout, conditions with high global prevalence. In previous work, we identified a uric acid (UA)-inducible gene cluster in Clostridium sporogenes that is required for converting uric acid to short-chain fatty acids (SCFAs)1. When these genes were disrupted, SCFA production was blocked, yet a substantial amount of uric acid was still consumed. This suggested the possibility that pathway intermediates accumulate in the mutants and could be identified by untargeted LC-MS.
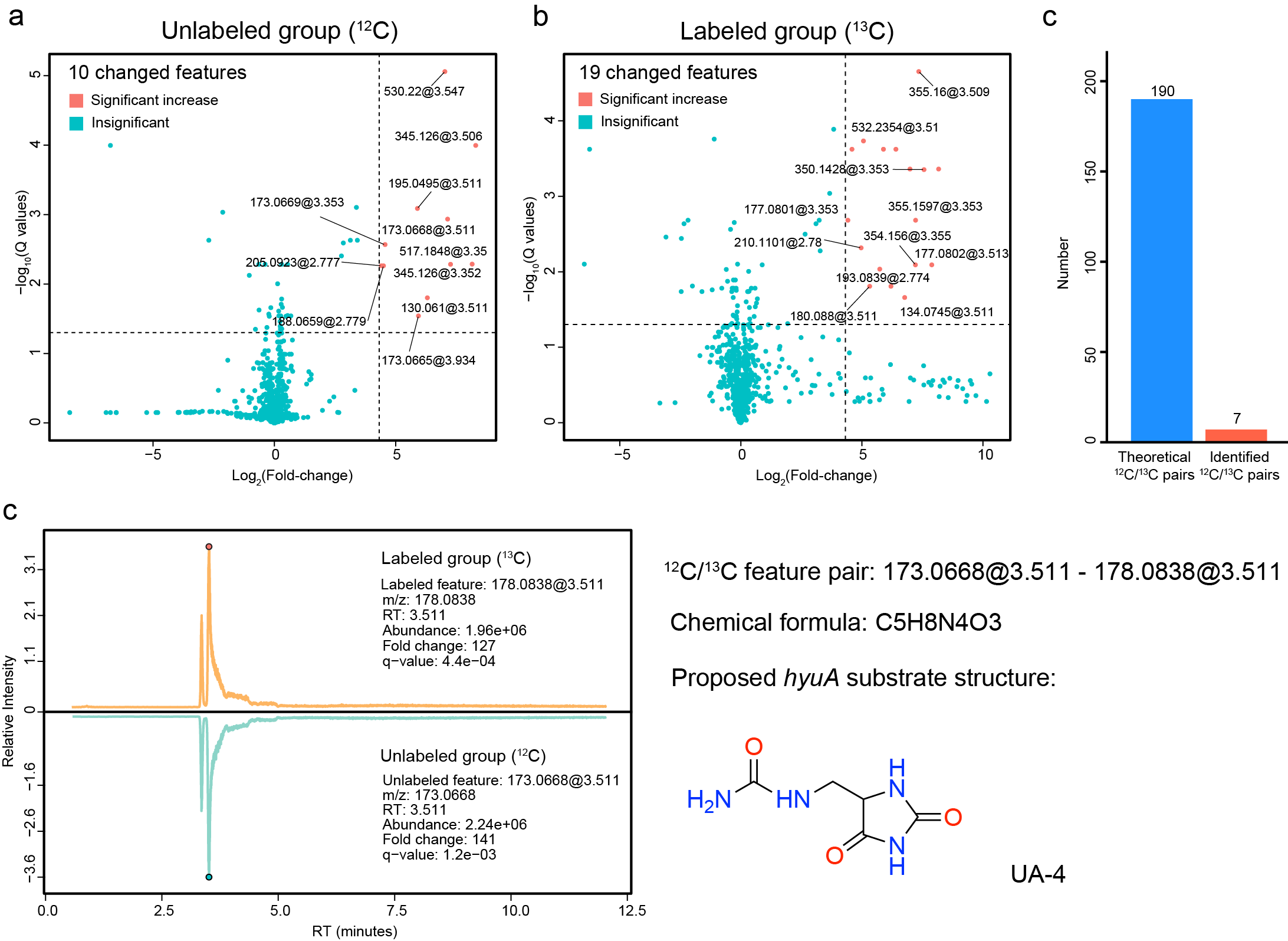
To investigate this, we developed a stable isotope tracing (STI) metabolomic experiment to detect unknown metabolites produced from uric acid in the anaerobic mutant cultures2. Specifically, for the UA catabolism gene cluster, we curated the 6 mutant strains for each gene. We cultured wild-type and mutant strains of C. sporogenes in the presence of either unlabeled uric acid or its [13C5]-labeled isotopologue. Here, the study of the hyuA gene was used as an example (Figure fig-figure4-1).
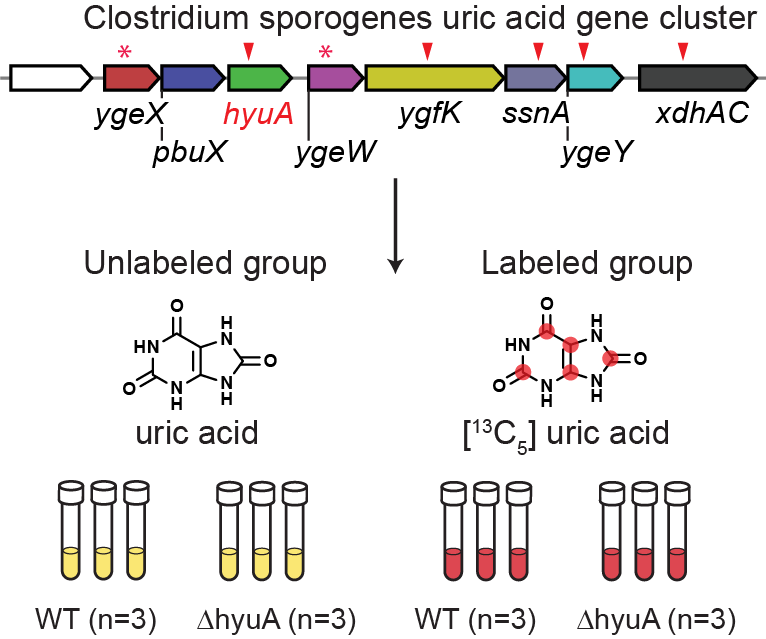
The reasonable substrate candidates have a series of characteristics: (i) were elevated in mutant vs. wild-type cultures, (ii) increased in intensity over culture age, and (iii) contained retention-time matched isotopologs when mutants were cultured with [13C5]-labeled uric acid. We processed these STI metabolomics data using IsoPairFinder. Take the hyuA as an example, compared to 190 possible pairs in the unlabeled and labeled groups, IsoPairFinder could significantly narrow the candidate pairs to 7. The final 12C/13C feature pair could be further quickly determined, which further helps propose its possible structure (Figure fig-figure4-2).